Kugako Sugimoto, NOST Tokyo
Origineel gepubliceerd op de site van Rijksdienst voor Ondernemend Nederland
Sammenvatting
Personalized medicine heeft als doel het effect van medicijnen te maximaliseren en tegelijkertijd de bijverschijnselen te minimaliseren door het gebruik van genetische informatie en de fysiologische conditie en ziektegeschiedenis van de patienten. Het verzamelen van genetische informatie is een belangrijk onderdeel van de ontwikkeling van personalized medicine. Japan heeft deelgenomen aan internationale genoomprojecten, technologieontwikkeling en methodes om personalized medicine te realiseren zoals het Human Genome Project (HGP). Bovendien heeft Japan geprobeerd biomakers te ontdekken, essentiёle hulpmiddelen voor personalized medicine. Echter, de verwezenlijking van personalized medicine lijkt nog geen volledig succes te zijn met inbegrip van het effectieve gebruik van pharmacogenomics. Biobanken om de verwezenlijking van personalized medicine te ondersteunen beginnen in aantal toe te nemen.
Development of personalized medicine in Japan
Summary
Personalized medicine aims to maximize the effect of the medicine and to minimize the side effect at the same time by using the genetic information and physiological/disease condition of the patients. Collection of the genetic information is an essential part for the development of personalized medicine. Japan has participated in international projects for genomes, technology development, and methods to realize personalized medicine such as Human Genome Project (HGP). In addition, Japan has tried to discover biomarkers, essential tools for personalized medicine. However, realization of personal medicine seems not to be fully successful including the effective use of pharmacogenomics. Biobanks to support realization of personalized medicine started to increase the numbers.
Details
Personalized medicine aims to find the best remedy for a patient by knowing his or her genetic information, physiological and disease conditions by using biomarkers in addition to the conventional medical information. This approach reduces the unwanted side effects and maximizes the effect of medicine. Personalized medicine is also beneficial for doctors, pharmaceutical industry, and governments. For doctors, selection of remedies would be based on scientific evidence avoiding the trial and error approach of medicine. For the pharmaceutical industry, the cost and time of drug creation would be decreased. For governments, medical expenditure would be reduced. However, there is still a long way to go before personalized medicine is globally available.
International project
It is difficult for one country alone to conduct the research to create personalized medicine. A person’s genetic information is one of the basic ingredients needed for development of personalized medicine. At the start of mapping the human genome, the process requires cutting-edge technology, information, a lot of money and a huge amount of work. Reading a human genome is estimated to have cost 3 billion dollars and taking 15 years. International projects have split the burden of money, labor and share the output. Japan has participated in several of these international projects hoping to create a road to personalized medicine.
Human Genome Project
Human Genome Project sequenced the human genome completely in 2003. From Japan, the RIKEN team previously known as Genomic Sciences Center and Department of Molecular Biology of Keio University School of Medicine participated in this project. Japan mapped 6% of the whole genome, while US covered 58%. RIKEN did one part of chromosome 11, 18, and 21 and Keio University read one part of chromosome 2, 6, 8, 21, and 22. This project provided the most basic information of genomic DNA to understand a human on the molecular level.
International HapMap Project
The International HapMap Project develops a haplotype map of the human genome. This map describes the common patterns of human DNA sequence variation. The Japanese Ministry of Education, Culture, Sports, Science and Technology funded participation of Japanese research groups. RIKEN and the University of Tokyo covered 24.3% of the genome by genotyping1 chromosomes 5, 11, 14, 15, 16, 17, and 19. The DNA samples for the HapMap included the Japanese people together with the Yoruba of Nigeria, Han Chinese, and U.S residents with ancestry from Northern and Western Europe.
Encyclopedia of the Human DNA Elements (ENCODE) Project
The ENCODE project identifies all functional elements in the human genome sequence. RIKEN participated in this project together other U.S., Spain, U.K., and Singapore. RIKEN used own genome analysis method, called CAGE technology2 and contributed to the project.
International Cancer Genome Consortium (ICGC)
ICGC aims to elucidate the genomic changes in many forms of cancers and to make comprehensive catalogues of genomic abnormalities in tumors from 50 different cancer types and/sub types. Japan leads the study of liver cancer-hepatocellular carcinoma3 (virus associated). Japan tries to elucidate the comprehensive somatic changes in the genome (mutation, rearrangement and copy number change), the epigenome (methylation et cetera) and transcriptome of virus (both HBV and HCV)-associated hepatocellular carcinoma. Hidewaki Nakagawa of RIKEN and Tatsuhiro Shibata of National Cancer Center Research Institute are the principal investigators in this project.
International Human Epigenome Consortium (IHEC)
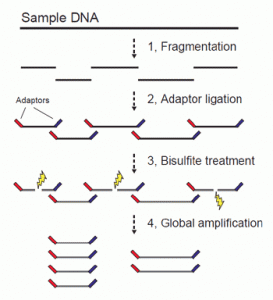
IHEC aims to decipher at least 1,000 epigenomes as well as creating of high-resolution maps of informative histone modifications, high-resolution DNA methylation maps, et cetera. Three Japanese teams are conducting research in these projects and cover different cells. One team focuses on the intestines such as liver, large intestine, stomach, and kidney. Japanese people have high ratio of stomach cancers and liver cancers, which are in need of effective remedies. This team uses an original analysis method called post-bisulfite adaptor-tagging (PBAT) and an algorithm to produce high quality data. In the sequence analysis by using a next generation sequencer, a segment of genomic DNA needs adaptors, constructed sequence, at the both ends of the DNA template. However, the conventional methods caused breakdown of template DNA and resulted in a low production of copied DNA. PBAT aims to decrease the breakdown of the template structure of DNA caused by bisulfite treatment to the template DNA with adaptor sequences at the both edges (Fig. 1). To decrease the chance of breakdown of the DNA template, adaptors are added after bisulfite treatment. By using PBAT method, preparation for a sequence analysis was possible from a small amount of DNA (125 pg) without the steps of polymerase chain reaction (PCR). In addition, it is possible to obtain data suitable for IHEC standard from only 5,000 egg cells of mice containing 30 nanograms of DNA.
Fig. 1 Unwanted breakdown of DNA sequence (IHEC Team Japan)
One team specializes in the cardiovascular system of endothelial cells such as coronary artery endothelial cells and aortic endothelial cells. The monoclonal antibody and oligoclonal antibody4 specific for histone modification are developed for ChIP-seq5. A ChIP-seq that is capable of handling a small amount of cells by using these antibodies is being developed now.
The third team focuses on reproduction organs such as placenta, uterus, and sperm cells. They reported high frequency of abnormality of DNA methylation of imprinting genes in sperms of hypospermatogenesis and high imprinting abnormality for invitro fertilization (IVF) children. The team is trying to obtain epigenome data of the endometrium and placenta to elucidate the cause of epigenome abnormalities.
Japan has participated in the international project for genomics, epigenomics, and other omics. However, Japan did not achieve good outcomes yet. For example, the human genome project did not promote basic research and technology development in Japan due to lack of budget and support from the government. To prevent wasting time and money on the research including preliminary research, long-term and wise strategy and policy of the Japanese government are needed.
Biobank
A biobank is an essential research source to elucidate the genetic causes needed to understand what environmental factors have an impact on health by genome analysis in addition to the conventional epidemiologic methods. A biobank is usually designed for cohort study. Participants are usually healthy persons. Since a biobank is not only for storage of biosamples, a lot of personal information such as family information is also stored. Proper management, security control of both information and biosamples are essential for operating biobanks.
Biobanks in Japan
Japan Public Health Center-based Prospective Study
The Japan Public Health Center-based prospective study started in 1990 and collected information from 140,000 people. Blood samples from 60,000 people were also stored. The aim of this study is to develop a policy and strategies against lifestyle related diseases. The National Cancer Research Institute has conducted this cohort study since 2010 and also started omics study by using the stored blood in 2011.
Biobank Japan
Biobank Japan is designed for a project called Realization of Order-made Medicine in 2003. This biobank is designed mainly for case control study6. However, it also has a cohort study aspect by collecting serum and clinical information in order to understand the process of development of disease on a molecular level. Samples and information of 47 diseases, 200,000 people, with 328,993 symptoms were collected by January 2012. Samples can be used by outside researchers beside University of Tokyo that manages the biobank. However, individual information is not supplied.
Cabinet Office Genome Cohort Research
This biobank is designed to store information of 100,000 people to integrate genome cohort studies and medical information to develop preventive medicine. This project is conducted by the National Cancer Research Center. The final goal is to obtain at least 300,000 people to be competitive at the international level with high quality and large data. To obtain large data, the methods to integrate results of previously existed cohort studies are being developed.
Tohoku Medical Mega Bank
This project is one of the reconstruction plans for the Tohoku area where a huge earthquake hit in 2011. The project aims to contribute to the reconstruction of the regional medical infrastructure and the construction of next generation medical system such as preventive medicine and personalized medicine by incorporating large genome cohort studies. Samples will be collected from the residents (150,000 people) of the Miyagi and Iwate prefectures in a period of 10 years. About 153,000,000 euro was already invested in the project by the end of March of 2013.
Pharmacogenomics (PGx)
Pharmacogenomics7 studies the role of genetics in drug response. Genetic variation of the patients such as SNP8 would respond differently, for example, in drug absorption, distribution, metabolism, and eliminations. Also, the receptor of a drug as a target molecule would be different among patients. For personalized medicine, PGx is an important tool especially in the development of a drug for a specific patient.
The Pharma SNP Consortium
Japanese companies created the Pharma SNP Consortium in 2000. The goal was to realize personalized medicine. The purpose of the consortium was to conduct pharmacokinetic research on Japanese gene polymorphism9 for three years. Specific studies identified SNP in a pharmacokinetics-related gene, frequency of SNP emergence in the Japanese people, and analysis of the expression and function of mutation-type protein generated under the influence of SNP. For the first time blood samples were collected from more than 1,000 Japanese people for a research purpose. Japanese companies invested about 7,200,000 euro in this project. Unfortunately, the data collected in this project was not used for drug development by Japanese companies. Technology and data at that time were not sophisticated enough to make use of it for real drug creation.
Japan PGx Data Science Consortium
The Japan PGx Data Science Consortium (JPDSC) was founded in 2009 by 6 Japanese leading pharmaceutical companies such as Astellas Pharma, Otsuka Pharmaceutical, Daiichi Sankyo, Taisho Pharmaceutical, Takeda Pharmaceutical, and Mitsubishi Tanabe Pharmaceutical. This consortium aims to build a DNA database for the drug creation for the Japanese people to check adverse reactions, efficacy and safety of drugs. In the first phase, 1,000 control samples have been genotyped.
Several studies used this DNA database and some of the results were already published. In 2011, a research team of National Institute of Health Sciences identified a useful biomarker for allopurinol-related Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN) in Japanese people. SJS/TEN are severe and coetaneous adverse drug reactions. 21 polymorphisms on chromosome 6 were significantly associated with allopurinol-related SYS/TEN. Strong association with allopurinol-induced SYS/TEN at rs9263726 in PSORS1C1, was found as well as at rw27334583 in BAT1, rs3094011 in HCP5 and GA005234 in MICC. Authors concluded that rs9263726 would be a good biomarker for allopurinol-related SJS/TEN in Japanese because of easy handling.
Currently, not only genomic approach but also other omics (metabolomics and epigenomics) and biosystematic approaches are used to realize personalized medicine. However in Japan, realization of personalized medicine seems to have some time to go. For the Japanese market, reliable and large biobanks that have at least 100,000 Japanese samples may help support to establish evidence. In addition, international cooperation on research is important and can be achieved by sharing data, evidences, and technologies to realize personalized medicine. Moreover, continuous effort of the government and companies for the large projects associated with strategic budgeting is important.
Sources
Post-bisulfite adaptor-tagging (PBAT) method (in Japanese)
Tohkin et al. Pharmacogenomics Journal (2013) 13, 60-69,
Research Paper #56 (Office of Pharmaceutical Industry Research, Akira Nagumo) (in Japanese)
International Human Epigenome Consortium, Team Japan
Liou et al. Drug Metab. Pharmacokinet(2012) 27(1), 2080
1 genotyping (Wikipedia)
3 hepatocellular carcinoma (Wikipedia)
5 ChIP-seq (Wikipedia)
8 SNP
9 polymorphism (Wikipedia)
Streamer
Japan has made an effort to realize personalized medicine by building biobanks and inventing original research methods.